Revisiting Cheminformatics and Mechanisms of Action of Chloroquine and Hy- droxychloroquine in Targeting Covid-19
Received Date: July 29, 2020 Accepted Date:August 08, 2020 Published Date: August 12, 2020
doi: 10.17303/jbcg.2020.3.101
Citation:Fidelis Toloyi Ndombera (2020) Revisiting Cheminformatics and Mechanisms of Action of Chloroquine and Hydroxychloroquine in Targeting Covid-19. J Bioinfo Comp Genom 3:1-11.
Abstract
COVID-19 is a global pandemic with adverse socioeconomic effects that continue to pose a real risk to human survival. The ever-increasing infections are projected to over 15 million infections and 1 million deaths by 2021. There are urgency and multiple efforts to find a cure, vaccines, and therapies to slow infection and Covid-19 related mortality. These efforts include research in repurposed and off-label use of drugs of which Chloroquine (CQ) and hydroxychloroquine (HCQ) have shown some promise. (CQ) therapy is among the list of drugs in the World Health Organization (WHO) SOLIDARITY study. Chloroquine has been approved by Chinese, South Korean, and Italian health authorities for the experimental treatment of COVID-19. Importantly, multiple studies and reports highlight the benefits of CQ and HCQ to Covid-19 patients. In this review, we draw a nexus of the genetics, biology, and pathology of Covid-19 to its cheminformatic features to help researchers, physicians and the public analyze potential mechanisms that make CQ and HCQ beneficial for off-label use. Of note, structural modification of HCQ and CQ is only possible with informed consideration of physicochemical, ADME, and Toxicity profiles of analogues using cheminformatic tools such as the swiss ADME. Furthermore, cheminformatic and bioinformatic tools are valuable when research time, human subjects, and clinical research are out of reach in a pandemic. This review endeavors to fill these gaps in pursuit of a Covid-19 therapy by revisiting the cheminformatics and mechanisms of action of Chloroquine and hydroxychloroquine.
Keywords: Covid-19; Chloroquine; Hydroxychloroquine; cheminformatics
Introduction
In December 2019, a novel virus causing pneumonia-like symptoms was reported in Wuhan, the capital of Hubei province, China [1-3]. Since that time, the disease has spread rapidly across the globe. The etiological agent is SARS- CoV-2, a new coronavirus, named due to the similarity of symptoms caused by SARS (severe acute respiratory syndrome) [4, 5]. The number of reported cases of COVID-19 infection may rise to over 15 million in early 2021 and mortalities continue to rise, raising concern about the likelihood of successful containment. Successful containment is hinged on the efficacy of candidate drugs against Covid-19 that include Chloroquine (CQ) and hydroxychloroquine (HCQ) [6-10]. Off-label use of CQ and HCQ requires an in-depth understanding of the molecular biology of SARS - CoV -2 in deciphering therapeutic mechanisms of potential drugs [11]. Towards this end, reviewing potential mechanisms of CQ and HCQ as well as pathological insights is crucial in understanding cheminformatics and bioinformatics of potential drugs and is beneficial in repurposing drugs that target Covid-19.
Classification
SARS - CoV -2 is a member of the coronaviridae family. These novel viruses belong to the order Nidovirales, subfamily orthocoronavirinae that is further classified into four genera; Betacoronavirus, Alphacoronavirus, Gamacoronavirus, and Deltacoronavirus [3]. The major sources of Alpha and Betacoronaviruses are bats, while Betacoronavirus and Gamacoronavirus originate from birds and swine [4,5,6]. Evidence from molecular analyses shows that SARS- CoV -2 is a novelBetacoronavirus [4] which is a member of subgenus sarbecovirus [2]. Two strains of the virus have caused outbreaks of severe respiratory diseases in humans: severe acute respiratory syndrome coronavirus (SARSCoV or SARS-CoV-1), which caused the 2002–2004 outbreak of SARS, and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is responsible for the 2019–20 Covid-19 pandemic.
Genetic Structure and Molecular biology
Coronavirus (nCoVs) including Covid-19 are enveloped, positive-stranded RNA viruses with nucleocapsid. nCoVs are round and sometimes pleiomorphic with about125nm diameter [12]. The single-stranded RNA(+ssRNA) genome of Covid-19 is about 29891 nucleotides in size with a G+C content of about 40%, encoding 9860 amino acids. The Covid-19 genome contains two flanking untranslated regions (UTRs) and a single long open reading frame encoding a polyprotein. The 2019-nCoV genome is arranged in the order of 5′-replicase (orf1/ab)-structural proteins [Spike (S)-Envelope (E)-Membrane (M)-Nucleocapsid (N)]−3′ [13]. Overall, the genomic structure is a 5′-cap structure and 3′poly-A tail [14]. Starting from the viral RNA, the synthesis of polyprotein 1a/1ab (pp1a/pp1ab) in the host is realized. The transcription works through the replication-transcription complex (RTC) organized in double-membrane vesicles and via the synthesis of sub-genomic RNA (sgRNAs) sequences. Notably, transcription termination occurs at transcription regulatory sequences, located between certain open reading frames (ORFs) that work as templates to produce sub-genomic mRNAs. In the atypical CoV genome, at least six ORFs can be present. Among these, a frame shift between ORF1a and ORF1b guides the production of both pp1a and pp1ab polypeptides that are processed by virally encoded chymotrypsin-like protease (3CLpro) or main protease (Mpro). Of note, the structural genes have been shown to interact with accessory genes such as 3a/b and 4a/b. and hemagglutinin esterase gene (HE). The SARS-CoV-2 genome is arranged similarly, although it lacks the HE gene, which is unique in some Betacoronaviruses [15]. Other ORF sencode for structural proteins, including spike, membrane, envelope, and nucleocapsid proteins. Additionally, papain-like proteases are required for producing non-structural proteins (nsps). Non-structural proteins encoded by the polyprotein form a replication transcription complex (RTC) in a double-membrane vesicle (DMV) [16].
The four major structural proteins coded for by SARSCoV-2 are; the Membrane (M), Spike (S), Envelope (E), and Nucleocapsid (N)[16-18]. The Spike protein is a class 1 viral trans membrane protein varying in size from 1160nm to 1400nm depending on the type of host. It occupies the virion surface in a trimer like fashion giving it a solar crown appearance [19]. The S protein interacts with the cell receptors of various hosts, facilitating the entry of the virus into a cell [19]. Furthermore, it plays a vital role in eliciting the host immune response and determination of host range and tissue tropism [19]. Among the corona viruses, the ectodomain region of the S protein is comparable in the organization. The S1 domain facilitates receptor binding, while the S2 domain is involved infusion [19]. Moreover, the S1 domain is divided into the N and C terminal domains, which are essential in receptor binding with the latter containing the receptor-binding motif (RBM) [15]. The trimeric S1 domain in the spike protein sits on top of the trimeric S2 stalk. Current research has identified 27 amino acid substitutions in the S protein of SARS-C0V- 2 within a 1273 length amino acid stretch, with six of these substitutions situated in the RBD (aa 357-528) and four in the receptor-binding motif of the CTD of S1 domain [16].
The Matrix protein gives a definite shape to the virus envelope and is also the most abundant protein. This protein is mainly involved in viral assembly [17]. The M protein is the most dynamic protein among corona viruses regarding the amino acid composition. The three trans membrane domains in the matrix protein are flanked by a short amino acid terminal inside the virion and a long carboxy chain outside the virion. Overall, the M-M interaction maintains the viral scaffold [16]. Presently no studies indicate any amino acid substitutions in SARS-CoV 2 M protein.
The E (Envelope) protein is an integral protein involved in pathogenesis, viral assembly, and release of the viral particles [17]. It is the smallest of the structural genes in SARS-CoV2, and the protein also acts as an ion channel. Inactivation of the E protein has been proven to alter virulence in corona viruses, as it changes the viral morphology and tropism. The E protein possesses a short hydrophilic amino-terminal domain (7-12 amino acids), a large hydrophobic trans membrane domain (25 amino acids), and a long hydrophilic C-terminal domain[20]. Generally, the E protein among corona viruses is conserved.
The N protein constitutes the only protein present in the nucleocapsid [17]. The N protein facilitates matrix protein interaction during assembly and has also been shown to increase the efficiency of transcription [17] [24,25]. The three highly conserved regions in the N protein are N terminal domain, RNA binding domain (linker region), and the C terminal domain. However, the NTD is the most diverse in both sequence and length [26]. The RNA binding domain is involved in cell signaling, and it also serves as an antagonist for interferon and RNA interference hence modulating antiviral response [27, 28] Present research has identified five mutations in the N protein of SARSCoV-2; two occurring in the intrinsically dispersed region (IDR, pos 25 &26), and one each in the CTD (position 344), NTD (pos 103) and LKR (pos 217).
Evolution
Sequence analysis of SARS-CoV-2 genome isolated from patients has revealed a 99.9% sequence identity, suggesting a very recent host shift to humans [1,2,3]. Various studies indicate that coronaviruses are evolutionary shaped and hosted by bats, and most coronaviruses in human hosts are derived from bat reservoirs[3,21]. Recent studies have confirmed a genetic resemblance between the 2019 novel virus and betacoronavirus from the sarbecovirus genus[21].
Population genetics analyses suggest that SARS-COV-2 viruses evolved into two major types, Land S, which are defined by two SNPs, position 8782 and 28144 [39]. Although L is more prevalent (70%) than S (30%), evolutionary analysis suggests that S is the more ancient type, and L is the more aggressive and infectious type. Recent molecular studies show that the divergence of SARS-C0V-2 and other coronaviruses had genomic nucleotide variability of 4% [39]. The analyses of divergence at the neutral sites between these viruses were found to be 17%, suggesting a more significant divergence than previously estimated. Lastly, this study suggested new differences in functional sites in the receptor-binding site of the S protein in SARS-CoV-2 that may have arisen due to natural selection and mutations besides recombination.
Pathological insights of Covid-19 patients treated with Chloroquine and hydroxychloroquine
The infection of host cells by SARS-CoV-2 is mediated by the S1 domain of the spike protein binding to angiotensin-converting enzyme 2 (ACE2) receptor, with the S2 domain facilitating fusing of the virus with the host cell membrane [22]. The SARS-CoV-2 virus is most likely initially taken up by nasopharyngeal mucosal cells before migrating into endothelial cells of the lung alveoli and into the bloodstream [23]. It then infects organs with cells bearing ACE2 receptors such as endothelial cells of the vasculature, myocardium, kidney, intestine, and brain. The onset of Covid-19 disease is characterized by fever, myalgia, cough, and dyspnea, and sometimes headache, diarrhea, nausea, and vomiting [24], symptoms that overlap with other viral syndromes. Covid-19 infection can result in severe illness, manifesting as systemic inflammatory response syndrome, acute respiratory disease syndrome (ARDS), [25] shock, and multiple organ failure in some cases [26-29]. The risk for severe Covid-19 disease includes old age and comorbidities such as cardiovascular disease, diabetes, and other respiratory diseases, although young and healthy individuals have also gone on to develop severe disease and or even death [30]. Several common laboratory abnormalities have been noted in Covid-19 patients. The most notable include increased inflammatory markers such as C-reactive protein, D-dimers, ferritin, and interleukin-6 (IL6) as well as elevated lactate dehydrogenase and lymphopenia [31]. Some of these parameters have also been found to be associated with disease severity, risk of requiring mechanical ventilation, intensive care unit [ICU] admission, or death. They include elevated D-dimers and thrombocytopenia [32]. Also implicated in the inflammation characteristic of severely ill Covid-19 patients is a 'cytokine storm' mainly of IL-6, IL-1β, and TNF-α [33].
There are several other hemostatic parameters in addition to D-dimer that are linked with Covid-19 disease severity, and which together point to some forms of coagulopathy that may predispose to thrombotic events [32, 34]. In a study of 183 COVID-9 patients, some hemostatic parameters were elevated in the 21 (11.5%) patients who died compared to those who survived. These include elevated D-dimer and fibrin degradation products (FDPs), prothrombin time (PT) elongation by 14% and 71% of those who died fulfilled the International Society on Thrombosis and Haemostasis (ISTH) criteria for disseminated intravascular coagulation (DIC) compared with only 0.6% among survivors [35]. Complications arising from DIC in Covid-19 include increased thrombin generation and micro thrombi with secondary parenchymal bleeding through endothelial leakage [36]. There may also be venous thromboembolism caused because of the immunological activation of thrombin derived from platelets or plasma [37]. Fatal pulmonary embolism may be the most frequent consequence of DIC, with this being a subject of ongoing Covid-19 pathology examinations [37]. Two possible pathological coagulation processes have been proposed in the clinical manifestations in critically ill Covid-19 patients. First, there is a local direct and endothelial injury resulting in thrombi formation and angiopathy in the lungs and other organs [38]. Secondly, in the systemic circulation, there is possible large vessel thrombosis and thromboembolic sequelae, all due to hypercoagulability, reported in 20-30% of patients admitted in ICU [39]. The ongoing pathological intervention of Covid-19 with CQ and HCQ is likely to consider the foregoing findings.
Chloroquine and hydroxychloroquine COVID-19 Therapy
Currently, many trials have been designed to determine an effective therapeutic regimen for COVID-19 [40]. Of the target regimens, chloroquine (CQ) therapy is also being considered and is among the list of drugs in the World Health Organization (WHO) SOLIDARITY study. Chloroquine has been approved by Chinese, South Korean, and Italian health authorities for the experimental treatment of COVID-19. On March the 28th day of 2020, the FDA authorized and later revoked the use of CQ and HCQ under an Emergency Use Authorization (EUA). On April 1st day of 2020, the European Medicines Agency (EMA) issued guidance that CQ and HCQ are only to be used in clinical trials or emergency use programs. Early clinical trials in China have shown chloroquine phosphate, an aminoquinoline used in malaria treatment, to be effective against COVID-19 [41] at a dose of 500 mg/day. In early in vitro studies, CQ blocked COVID-19 infection at low-micromolar concentration, with a half-maximal effective concentration (EC50) of 1.13 μM and a half-cytotoxic concentration (CC50) greater than 100 μM [42]. CQ is a 9-aminoquinoline that has been known since 1934 and is specifically synthesized to be used as an antimalarial agent. The parent compound, quinine, was isolated in the late 19th century from the bark of the tropical cinchona tree [43]. Unfortunately, CQ is being gradually dismissed from antimalarial therapy and prophylaxis, due to the continuous emergence of chloroquine-resistant Plasmodium falciparum strains [44]. However, the tolerability, low cost, toxicity, and immunomodulatory properties of CQ and HCQ are associated with biochemical effects that suggest a potential use in viral infections, some of whose symptoms may result from the inflammatory response [45]. Importantly, cheminformatics reveals unique structural features that enhance the drug properties of CQ and HCQ.
Chemical and Physicochemical properties of Chloroquine and hydroxychloroquine
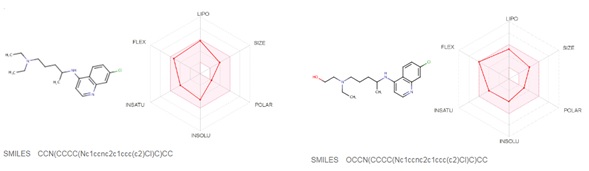
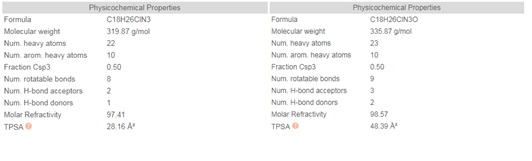
Chloroquine (7-chloro-4-(4-diethylamino-1-methyl butyl amino) quinoline) is prepared by the condensation of 4-7-dichloroquinoline with 1-diethylamino-4-amino pentane. Chloroquine (CQ) is a white to yellow, odorless crystalline powder with a bitter taste with a melting point between 87 to 92°C. CQ is very slightly soluble in water but is soluble in chloroform, ether, and dilute acids. A cheminformatic analysis of the physicochemical properties of CQ further reveals pharmacophore features that influence its behavior as a potent drug that can target Covid-19. The swissADME [46] Bioavailability Radar provides a graphical snapshot of the drug parameters of an orally available bioactive drug. The Bioavailability Radar plot is presented as a hexagon (Figure1) with each of the vertices representing a physicochemical parameter that define a bioavailable drug. The pink area within the hexagon represents the optimal range for each property (lipophilicity: XLOGP3 between -0.7 and +5.0, size: MW between 150 and 500 g/mol, polarity: TPSA between 20 and 130 Å2 , solubility: log S not higher than 6, saturation: the fraction of carbons in the sp3 hybridization not less than 0.25, and flexibility: no more than nine rotatable bonds). Of note, are the physicochemical properties of CQ and HCQ (Figure 2) that make them readily bioavailable drugs. Importantly HCQ with a terminal hydroxyl group that increases its topological polar surface area (TPSA) to 48.39Å2 compared to CQ’s 28.16Å2 . TPSA is vital in the prediction of biological barrier crossing of a drug such as in absorption and brain access. TPSA is a fragmental technique that considers polar surface atoms that includes sulfur and phosphorous to estimate the total polarity of a molecule. Additionally, HCQ becomes slightly more water-soluble and less lipophilic than CQ as depicted by the water solubility (Figure 3) and lipophilicity metrics (Figure 4). The partition coefficient between n-octanol and water (log Po/w) is the classical descriptor for lipophilicity [46].
In silico pharmacokinetics of CQ and HCQ
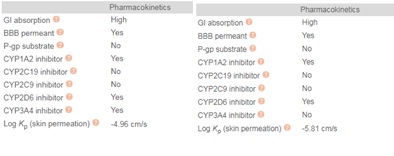
Metabolism of HCQ involves CYP3A4 and CYP2C3-driven dealkylation to form desethyl hydroxychloroquine, desethylchloroquine, and bisdesethyl chloroquine. In addition, swissADME metrics reveal that HCQ does not inhibit CYP3A4 while CQ could be an inhibitor (Figure 5). Additionally, both CQ and HCQ do not inhibit CYP2C19 and CYP2C9. CYP2C19 is a liver enzyme and a member of the CYP2C subfamily of the cytochrome P450 mixed-function oxidase system involved in the metabolism of xenobiotics, including many proton pump inhibitors and antiepileptics. Inhibition metrics of other cytochrome p-450 enzymes are as in figure 5 below.
ADME of Chloroquine and hydroxychloroquine
Chloroquine's (CQ) absorption is rapid and is widely distributed in body tissues. It has a very high volume of distribution, as it diffuses into the body's adipose tissue. CQ’s protein binding is high, with a half-life of around 50 days [47]. CQ is metabolized to desethylchloroquine partially by the liver and more than 50% excreted as unchanged drug in urine, where acidification of urine increases its elimination. Accumulation of the drug may result in deposits that can lead to blurred vision and blindness. Also, its related quinines have been associated with cases of retinal toxicity, particularly when provided at higher doses for longer times.
HCQ is administered orally, in doses ranging from 100 to 1200 mg daily, and is absorbed within 4 hours. It is approximately 50% bound to plasma protein in the blood. HCQ blood concentration peaks after the absorption phase and falls quickly due to rapid partitioning into organs. Accumulation in lysosomes appears to drive the large volume of distribution in plasma. Excretion takes place mainly in the kidneys, accounting for about a quarter of HCQ total blood clearance, with liver clearance assumed to account for the rest [48].
Drug likeness of Chloroquine and hydroxychloroquine
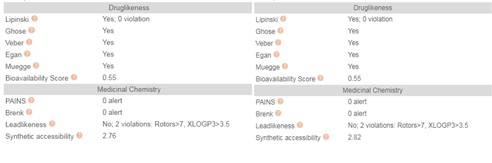
The phrase “drug-like” is defined as those compounds that have sufficiently acceptable ADME and toxicity properties to survive through the completion of Phase I clinical trials [49]. Drug-likeness is a complex balance of molecular properties and structural features that determine whether an unknown molecule is like the known drugs. These molecular properties include hydrophobicity, electronic distribution, and hydrogen bonding characteristics, molecule size, and flexibility. SwissADME has computational filters that include Ghose [50], Egan [51], Veber [52], Muegee [53], and Lipinski rules used by leading pharmaceutical companies and chemoinformatics to evaluate the drug-likeness of small molecules.
The Ghose filter quantitatively characterizes the structure of a molecule based on computed physicochemical property profiles that include log P, molar refractivity (MR), molecular weight (MW), and the number of atoms. In addition, the Ghose filter includes a qualitative characterization based on the presence of functional groups and important substructures. The qualifying range of calculated log P (ClogP) is between -0.4 and 5.6. For MW, the qualifying range is between 160 and 480. For MR, the qualifying range is between 40 and 130, and for the total number of atoms, the qualifying range is between 20 and 70 atoms in a small molecule. Notably, both CQ and HCQ meet all these Ghose criteria (Figure7).
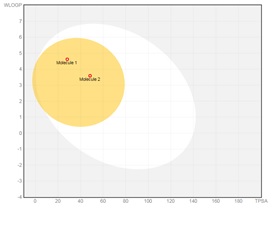
Egan (Pharmacia) filter provides a prediction of drug absorption based on physical processes involved in membrane permeability of a small molecule. Importantly, the Egan computational model for human passive intestinal absorption (HIA) of small molecule accounts for active transport and efflux mechanisms and is therefore robust in predicting the absorption of drugs. Both CQ and HCQ pass the Egan filter largely due to their polar surface area, and a number of hydrogen acceptors (Figure 1 & 2) that influence their hydrophilicity and hydrophobicity (Figure 3 & 4). This is because the descriptors in the Egan model are polar surface area (PSA) and AlogP98v with the exclusion of redundant descriptors such as MW. PSA is a reference point for AlogP98. AlogP98 descriptor is a ratio of lipophilicity to hydrophilicity which contains no information on the absolute measure of either factor.
Veber (GSK filter) model characterizes molecules as drug-like if they have ten or fewer rotatable bonds and a PSA equal to or less than 140 Å2 with 12 or fewer H-bond donors and acceptors (Figure 2). Molecules with these properties have a high probability of good oral bioavailability. CQ and HCQ molecules met Veber criteria. In addition, CQ and HCQ met the Muegge rules of Druglikeness.
Muegge (Bayer filter) model is a database-independent pharmacophore point filter that discriminates between drug-like and nondrug-like chemical matter. It is based on the observation that non-drugs are often less functionalized. Four functional motifs are defined to be important in drug-like molecules and include ketone, hydroxyl, sulfonyl, and amine groups. The occurrence of these functional motifs guarantees hydrogen-bonding capabilities that are essential for specific drug interactions with its targets. CQ and HCQ have an aminoquinoline pharmacophore. In addition, HCQ has an N-hydroxy-ethyl side chain in place of the N-diethyl group of CQ thus providing essential hydrogen bond acceptors and donors with their targets (Figure 2). These functional groups can be combined with what the Muegge model [54] refers to as pharmacophore points. The pharmacophore points include amine, amide, alcohol, ketone, sulfone, sulfonamide, carboxylic acid, carbamate, guanidine, amidine, urea, and ester functional groups.
Insights to mechanisms of action of Chloroquine and hydroxychloroquine
Chloroquine (CQ) and hydroxychloroquine (HCQ) are weak bases that affect acid vesicles leading to the dysfunction of several enzymes. CQ is also a lysosomotropic agent, meaning it accumulates preferentially in the lysosomes of cells in the body. Interference of lysosomal activity inhibits the function of lymphocytes and has immunomodulatory and anti-inflammatory effects [47].In vitro, CQ can destabilize lysosomal membranes and promote the release of lysosomal enzymes inside cells [47]. The pKa for the quinoline nitrogen of chloroquine is 8.5, meaning it is about 10% deprotonated at physiological pH according to the Henderson-Hasselbalch equation. This decreases to about 0.2% at a lysosomal pH of 4.6. Since the deprotonated form is more membrane-permeable than the protonated form, a large amount accumulates in lysosomes.
Extracellularly, CQ and HCQ are present in a protonated form that and thus are unable to cross the plasma membrane. However, the non-protonated portion can enter the intracellular compartment and become protonated in an inversely proportional fashion to the internal pH. Accordingly, CQ and HCQ are concentrated within acidic organelles such as the endosome, Golgi vesicles, and the lysosomes, where the pH is low. This low pH in lysosomes is optimal for lysosomal enzymes involved in hydrolysis, and so by increasing the pH of endosomal compartments, CQ and HCQ disrupt the maturation of lysosomes and autophagosomes and inhibit antigen presentation along the lysosomal pathway[47].
CQ and HCQ also act by altering protein degradation pathways through acidic hydrolases in the lysosomes, macromolecule synthesis in the endosomes, and post-translational protein modification in the Golgi apparatus. Additionally, high CQ-mediated endosomal pH modulates iron metabolism and impairs the release of iron from ferrated transferrin, to lower the intracellular concentration of iron [55]. This decrease causes the dysfunction of several cellular enzymes with implications in DNA replication and gene expression [55]. Studies have also demonstrated that CQ confers broad-spectrum antiviral effects via this pH-lowering mechanism that affect viral-fusion processes. Moreover, CQ can alter the glycosylation of the cellular receptors of coronaviruses because it involves proteases and glycosyl-transferases, some of which require a low pH.
Hydroxychloroquine (HCQ), a less toxic aminoquinoline, has an N-hydroxy-ethyl side chain in place of the N-diethyl group of CQ. HCQ has a modulating effect on activated immune cells, downregulates the expression of Toll-like receptors (TLRs) and TLR-mediated signal transduction, and decreases the production of interleukin-6 [47]. Changes in endosomal pH can interfere with TLR9 and TLR7 processing, and thus CQ and HCQ prevent TLR activation upon extracellular stimuli by mediating changes in the local pH. Of note, CQ can bind to nucleic acids and thus prevents the activation of endosomal TLRs [56].
HCQ is preferred over CQ for its lower ocular toxicity. Furthermore, in patients with COVID-19, CQ interacted with lopinavir/ritonavir, resulting in prolongation of the QT interval. Conversely, retinopathy is a dose-limiting adverse effect of hydroxychloroquine. However, a safe daily dose seems to correspond to 6.5 mg/kg of the ideal body weight and 5.0 mg/kg of the actual body weight [47]. Of note, there are more CQ clinical data than those about HCQ as an antiviral agent.
HCQ is also a lysosomotropic autophagy inhibitor being used in many clinical trials, either alone or in combination with chemotherapy [48]. Mechanistically, HCQ being a weakly basic compound that basifies the highly acidic lysosome prevents the autophagosome-lysosome fusion step of autophagy. This mechanism drives its pharmacokinetics (PK), mainly through an ion-trap accumulation observed in acidic compartments of a cell, including lysosomes [48].
Some viruses enter their target cells by endocytosis. CQ inhibits different viruses that require a pH-dependent step for entry into cells. Of note, CQ reduces the secretion of proinflammatory cytokines such as TNF alpha [57, 58]. Furthermore, treatment with HCQ inhibits the production of TNF, IFNα, IL6, and CCL4 (also known as MIP1β) in pDC. In vitro, HCQ and CQ impede the production of IL-1, IL-6, TNF and IFNγ by mononuclear cells.
Conclusion
The use of CQ and HCQ in targeting Covid-19 relies on a thorough understanding of its genetic structure, molecular biology, evolution as well as known and predictable mechanisms of action of CQ and HCQ. Additionally, cheminformatic and bioinformatic analysis of CQ and HCQ is beneficial in repurposing these malarial drugs for Covid-19. Importantly known management of Covid-19 with CQ and HCQ provides useful insights and opens the opportunity for repurposing current drugs to mitigate the lethal effects of Covid-19.
- Velavan TP, and CG Meyer (2020) The COVID-19 epidemic. Tropical Medicine & International Health, 25: 278-280.
- Lu R, et al. (2020) Genomic characterization and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395: 565-574.
- Woo PCY, et al. (2012) Discovery of seven novel Mammalian and avian coronaviruses in the genus delta corona virus supports bat corona viruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. Journal of virology 86: 3995-4008.
- Lake MA (2020) What we know so far: COVID-19 current clinical knowledge and research. Clinical Medicine 20: 124- 127.
- Ren LL, et al. (2020) Identification of a novel coronavirus causing severe pneumonia in humans: a descriptive study. Chin Med J (Engl).
- McKee DL, et al. (2020) Candidate drugs against SARS-CoV-2 and COVID-19. Pharmacol Res 157: 104859.
- Gautret P, et al. (2020) Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents 105949.
- Pastick KA, et al. (2020) Review: Hydroxychloroquine and Chloroquine for Treatment of SARS-CoV-2 (COVID-19). Open Forum Infect Dis, 7: ofaa130.
- Touret F and X de Lamballerie (2020) Of chloroquine and COVID-19. Antiviral Res 177: 104762.
- Marmor MF (2020) COVID-19 and Chloroquine/Hydroxychloroquine: is there Ophthalmological Concern? Am J Ophthalmol 213: A3-a4.
- Schlagenhauf P, et al. (2020) Repurposing antimalarials and other drugs for COVID-19. Travel Med Infect Dis 34: 101658.
- Malik YA (2020) Properties of Coronavirus and SARSCoV-2. Malays J Pathol, 42: 3-11.
- Zhou P, et al. (2020) A pneumonia outbreak associated with a new corona virus of probable bat origin. Nature 579: 270-273.
- Li G, et al. (2020) Coronavirus infections and immune responses. J Med Virol 92: 424-432.
- Pastick KA, et al. (2020) Review: Hydroxychloroquine and Chloroquine for Treatment of SARS-CoV-2 (COVID-19). Open Forum Infectious Diseases 7.
- Chen Y, Q Liu, and D Guo (2020) Emerging coronaviruses: Genome structure, replication, and pathogenesis. J Med Virol 92: 418-423.
- Fehr AR, S Perlman (2015) Coronaviruses: an overview of their replication and pathogenesis. Methods in molecular biology (Clifton, N.J.)1282: 1-23.
- Hussain S, et al. (2005) Identification of novel subgenomic RNAs and noncanonical transcription initiation signals of severe acute respiratory syndrome coronavirus. Journal of virology79: 5288-5295.
- Masters PS (2006) The molecular biology of coronaviruses. Adv Virus Res 66: 193-292.
- Schoeman D and BC Fielding (2019) Coronavirus envelope protein: current knowledge. Virology journal16: 69-69.
- FanY, et al. (2019) Bat Coronaviruses in China. Viruses 11.
- Huang Q and A Herrmann (2020) Fast assessment of the human receptor-binding capability of 2019 novel coronavirus (2019-nCoV). bioRxiv 2020.02.01.930537.
- Harenberg J and E Favaloro (2020) COVID-19: progression of the disease and intravascular coagulation - present status and future perspectives. Clin Chem Lab Med 58: 1029-1036.
- Huang C, et al. (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395: 497- 506.
- Wu Z and JM McGoogan (2020) Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72 314 Cases From the Chinese Center for Disease Control and Prevention. Jama.
- Iba T and JH Levy (2019)Derangement of the endothelial glycocalyx in sepsis. J Thromb Haemost 17: 283-294.
- Du Y, et al. (2020) Clinical Features of 85 Fatal Cases of COVID-19 from Wuhan. A Retrospective Observational Study. Am J Respir Crit Care Med 201: 1372-1379.
- Su H, et al. (2020) Renal histopathological analysis of 26 postmortem findings of patients with COVID-19 in China. Kidney Int 98: 219-227.
- Varga Z, et al. (2020) Endothelial cell infection and endotheliitis in COVID-19. Lancet 395: 1417-1418.
- Wu C, et al. (2020) Risk Factors Associated With Acute Respiratory Distress Syndrome and Death in Patients With Coronavirus Disease 2019 Pneumonia in Wuhan, China. JAMA Internal Medicine.
- Wang D, et al. (2020) Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. Jama 323: 1061-1069.
- Lippi G, M Plebani and BM Henry (2020) Thrombocytopenia is associated with severe coronavirus disease 2019 (COVID-19) infections: A meta-analysis. Clin Chim Acta 506: 145-148.
- Chen G, et al. (2020) Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest 130: 2620-2629.
- Bikdeli B, et al. (2020) COVID-19 and Thrombotic or Thromboembolic Disease: Implications for Prevention, Antithrombotic Therapy, and Follow-Up. JACC State-of-the-Art Review 75: 2950-2973.
- Levi M, et al. (2009) Guidelines for the diagnosis and management of disseminated intravascular coagulation. British Committee for Standards in Haematology. Br J Haematol 145: 24-33.
- Bikdeli B, et al. (2020) COVID-19 and Thrombotic or Thromboembolic Disease: Implications for Prevention, Antithrombotic Therapy, and Follow-Up: JACC State-of-the-Art Review. J Am Coll Cardiol 75: 2950-2973.
- Tian S, et al. (2020) Pathological study of the 2019 novel coronavirus disease (COVID-19) through postmortem core biopsies. Mod Pathol 33: 1007-1014.
- Monteil V, et al. (2020) Inhibition of SARS-CoV-2 Infections in Engineered Human Tissues Using Clinical-Grade Soluble Human ACE2. Cell 181: 905-913.e7.
- Arjomandi Rad A, R Vardanyan and NR Tas (2020) Ibuprofen and thromboembolism in SARS-COV2. J Thromb Haemost.
- Touret F and X de Lamballerie (2020) Of chloroquine and COVID-19. Antiviral research 177: 104762-104762.
- Gao J, Z Tian and X Yang (2020) Breakthrough: Chloroquine phosphate has shown apparent efficacy in the treatment of COVID-19 associated pneumonia in clinical studies. Biosci Trends 14: 72-73.
- Wang M, et al. (2020) Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res 30: 269-271.
- Gachelin G, et al. (2017) Evaluating Cinchona bark and quinine for treating and preventing malaria. Journal of the Royal Society of Medicine 110: 31-40.
- Ocan M, et al. (2019) Persistence of chloroquine resistance alleles in malaria-endemic countries: a systematic review of burden and risk factors. Malar J 18: 76.
- Savarino A, et al. (2001) The anti-HIV-1 activity of chloroquine. J Clin Virol 20: 131-135.
- Daina A, O Michielin and V Zoete (2017) SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness, and medicinal chemistry friendliness of small molecules. Scientific Reports 7: 42717.
- Schrezenmeier E and T Dörner (2020) Mechanisms of action of hydroxychloroquine and chloroquine: implications for rheumatology. Nature Reviews Rheumatology 16: 155-166.
- Collins KP, KM Jackson and DL Gustafson (2018) Hydroxychloroquine: A Physiologically-Based Pharmacokinetic Model in the Context of Cancer-Related Autophagy Modulation. The Journal of pharmacology and experimental therapeutics 365: 447-459
- Lipinski CA (2000) Drug-like properties, and the causes of poor solubility and poor permeability. J Pharmacol Toxicol Methods 44: 235-249.
- Ghose AK, VN Viswanathan and JJ Wendoloski (1999) A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. Qualitative and quantitative characterization of known drug databases. J Comb Chem 1: 55-68.
- Egan WJ, KM Merz Jr, and JJ Baldwin (2000) Prediction of drug absorption using multivariate statistics. J Med Chem 43: 3867-3877.
- Veber DF, et al. (2002) Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem 45: 2615- 2623.
- Muegge I, SL Heald and D Brittelli (2001) Simple selection criteria for the drug-like chemical matter. J Med Chem 44: 1841- 1846.
- Muegge I (2003) Selection criteria for drug-like compounds. Medicinal Research Reviews 23: 302-321.
- Byrd TF and MA Horwitz (1991) Chloroquine inhibits the intracellular multiplication of Legionella pneumophila by limiting the availability of iron. A potential new mechanism for the therapeutic effect of chloroquine against intracellular pathogens. The Journal of clinical investigation 88: 351-357.
- Kužnik A, et al. (2011) Mechanism of Endosomal TLR Inhibition by Antimalarial Drugs and Imidazoquinolines. The Journal of Immunology 186: 4794-4804.
- Weber SM and SM Levitz (2000) Chloroquine interferes with lipopolysaccharide-induced TNF-alpha gene expression by a non-lysosomotropic mechanism. J Immunol 165: 1534-1540.
- Jeong JY, et al. (2002) Chloroquine decreases cell-surface expression of tumor necrosis factor receptors in human histiocytic U-937 cells. Immunology 105: 83-91.
 Figure 1
Figure 1
 Figure 2
Figure 2
 Figure 3
Figure 3
 Figure 4
Figure 4
 Figure 5
Figure 5
 Figure 6
Figure 6
 Figure 7
Figure 7