Human Leucocyte Antigen-G (HLA-G) 3’ UTR Genetic Polymorphisms associate with HIV-1 High-Risk Seronegative Status in the Amsterdam Cohorts Studies
Received Date: Septmber 19, 2020 Accepted Date: October 10, 2020 Published Date: October 12, 2020
doi: 10.17303/jaid.2020.7.104
Citation:Ahmed Alyami (2020) Human Leucocyte Antigen-G (HLA-G) 3’ UTR Genetic Polymorphisms associate with HIV-1 High-Risk Seronegative Status in the Amsterdam Cohorts Studies. J HIV AIDS Infect Dis 7: 1-12.
Abstract
HLA-G is a non-classical class I HLA molecule associated with the suppression of immune responses during infection. We investigated HLA-G 3’ UTR genetic polymorphisms and HLA-G protein expression in HIV-1 high-risk seronegative (HRSN) as well as infected individuals (HIV-1+). DNA samples from The Amsterdam Cohort Studies (ACS), comprising HIV-1+ MSM (Men who have Sex with Men) and IDU (Intravenous Drug Users), together with HRSN individuals were analyzed for HLA-G 3’ UTR (untranslated region) genotypes and serum soluble HLA-G (sHLA-G) plasma levels. HLA-G 3’ UTR haplotypes were determined through PCR amplification and subsequent DNA sequence analysis from the HIV-1 groups. sHLA-G concentrations were assayed through ELISA. Flow cytometry and real-time PCR technologies were used to identify whether leucocytes from healthy controls stimulated with HIV-1 pseudo-typed virus particles modulated HLA-G protein or mRNA expression levels, respectively. HLA-G 3’ UTR genetic polymorphisms within MSM and IDU HIV-1+ individuals were found to be similar to healthy controls whilst HRSN subjects showed a significantly higher proportion of specific 3’ UTR alleles previously associated with low levels of HLA-G expression: 3010C, 3027A, 3035T, 3142G and 14bpI (P< 0.01) and haplotypes associated with lower levels of expression, UTR2, 5 & 7, were significantly better represented in the HRSN group (p= 0.0217). No differences were found in sHLA-G expression between the variant HIV-1+ groups or controls or between individuals carrying variant HLA-G 3’ UTR genotypes. Exposure of leucocytes to HIV-1 pseudo-typed viral particles had no effect on modulating HLA-G protein or mRNA expression. These results indicate that HLA-G 3’ UTR genetic polymorphisms linked with low HLA-G protein expression associate with protection against HIV-1 transmission in the ACS.
Keywords: HIV; HLA-G; soluble HLA-G; CD4+ T cells; flow cytometry
Introduction
Human Immunodeficiency Virus type 1 (HIV-1) is a serious global health issue with approximately 2 million new infections reported annually. In 2014, an estimated 1.2 million people died from acquired immune deficiency syndrome (AIDS) or AIDS-related illnesses resulting from HIV-1 infections [17,18]. Both men who have sex with men (MSM) and injecting drug users (IDU) are at high risk for HIV-1 acquisition and continue to pose challenges for HIV-1prevention. MSM comprise one of the highest risk groups for HIV-1 infection worldwide [21].
HLA-G is a non-classical class I HLA molecule typically expressed on extravillous cytotrophoblasts and in low levels on some leucocyte subsets [36]. However, in addition to its protective role in the maintenance of pregnancy, it can also be involved in the evasion of immune responses against some infectious diseases or malignancies [39]. During viral infection, viruses can modulate the nonclassical HLA class I antigen HLA-G via diverse strategies such as disrupting immune-competent cell recognition. HLA-G cell surface expression was found to be significantly induced in H1N1 infection when compared to non-infected controls, suggesting that elevated cell surface expression may favour virus evasion from host immune responses [10]. Differences in HLA-G surface expression on monocytes, plasma soluble HLA-G (sHLA-G), IL-10, and IL-6 levels have been linked with Enterovirus 71 infection [56]. Similarly, HLA-G expression has been reported on hepatocytes and biliary epithelial cells within the livers of chronic hepatitis C patients [3]. Taken together, this suggests that viral infections can induce HLA-G expression, which could be a mechanism enabling viruses to subvert host immune defences.
In the context of HIV-1 infection, up-regulation of HLA-G cell surface expression on monocytes and on some T lymphocytes in HIV-1 positive individuals, irrespective of receiving antiretroviral therapy (ART) was reported [9, 29]. Increased expression of HLA-G at the fetomaternal interface was also noted in conjunction with HIV-1 infection [37]. Conversely, there are reports of ART inducing HLA-G expression on monocytes [12] and of HIV-1 down regulating HLA-G expression [14]. Furthermore, patients with the HLA-G*0501N allele, which does not encode for a functional HLA-G protein, were reported to be more resistant to HIV-1 infection [24, 35]. Therefore, understanding the relationship between HLA-G expression and HIV-1 infection is important not only to identify and develop new effective drug therapies and vaccines against HIV-1 but also to improve diagnostic predictions [17].
In healthy individuals, levels of serum sHLA-G may increase as a result of physiological or pathological conditions, such as pregnancy, acute rejection episodes after organ transplantation, autoimmune diseases, and some cancers. Interestingly, sHLA-G serum levels are elevated during some viral infections, including hepatitis B [47], and hepatitis C [54], influenza [10], cytomegalovirus [43], and HIV-1. Previously, the serum levels of sHLA-G were reported to be significantly elevated in HIV-infected subjects, and this was observed to influence disease progression and development of AIDS in addition to modulating the virologic and immunologic response to ART [18]. Several reports have linked sHLA-G levels to HIV-1 infection and virus susceptibility [15,20,23,25,40,41,49], with levels decreasing following initiation of ART [40,41].
Unlike classical class I MHC molecules, HLA-G has limited coding region genetic polymorphisms. However, there is a 3’ untranslated region (UTR) dimorphism of HLA-G dependent upon deletion or insertion of a 14bp sequence which affects levels of HLA-G expression [49]. This and two further 3’UTR dimorphic sites, +3142C/G and +3187G/A have been suggested to be associated with HLA-G mRNA expression regulation with the 14bp deletion, 3142C and 3187G alleles being associated with increased expression [22, 52, 55]. To date, 44 haplotypes of the HLA-G 3′ untranslated region have been identified which contain eight variable sites. UTRs 1-7 are the most frequent 3’UTR haplotypes, accounting for more than 96% of all the reported haplotypes worldwide [6, 31, 34], although further rarer haplotypes have been identified [31]. UTRs 2, 5 and 7 possess all alleles associated with high levels of HLA-G expression whereas UTR7 possesses all alleles associated with low levels of expression [8].
The aim here was to investigate the association of HLA-G genetic polymorphisms and expression levels between different clinical groups (HRSN versus HIV-1+) within the Amsterdam Cohorts Studies (ACS).
Materials and Methods
HIV patients and healthy controls
DNA and plasma samples from the time of diagnosis from various HIV-1 groups within the Amsterdam Cohort Studies (ACS) and healthy uninfected control individuals were utilized [12]. Further uninfected donors were obtained from the Institute of Infection and Global Health, the University of Liverpool for HLA-G in vitro expression analysis. This study has been conducted in accordance with the ethical principles set out in the declaration of Helsinki and was approved by the institutional review board of the Amsterdam University Medical Centers (location Academic Medical Center). Written informed consent was obtained from all participants.
HLA-G 3’UTR genetic polymorphism characterization
DNA samples from individuals within the different HIV-1 groupings and healthy controls were PCR amplified, sequenced, and analyzed for HLA-G genetic polymorphisms. Primers were designed via free commercial software (Oligo Perfect™ Designer, Thermo Fisher Scientific Inc. Web Tools). DNA was amplified with a set of two primers spanning the 3’UTR: 5’-GTG ATG GGC TGT TTA AAG TGT CAC C-3’, 5’-GGA AGG AAT GCA GTT CAG CAT GA-3’ (Sigma Aldrich). PCR reactions were carried out with standard reagents using a thermos-cycler (Techne Thermal Cycler, Keison International Ltd. UK) with the program set as follows: Initial denaturation at 94o C for 5min, followed by 30 cycles of denaturation at 94° C for the 30s, annealing at 64° C for 45s, an extension for 60s at 72° C and a final extension at 72° C for 10 min.
All the cleaned purified PCR products were sent off for Sanger sequencing using an ABI Big Dye sequencer (Source Biosciences Inc. UK). The reverse primer (3’ ACACGTGTACTGTGGAAAGTT ’5) was utilized for sequencing the DNA samples using the amplification process above. The results were reported electronically and the data were accessed with commercial software to manipulate and organize the data.
The sequences obtained were analysed manually for specific SNPs by observing the 14 bp INS/DEL (rs1704), +3003 C/T (rs1707), +3010 C/G (rs1710), +3027 A/C (rs17179101), +3035 C/T (rs17179108), +3142 C/G (rs1063320), +3187 A/G (rs9380142) and +3196 C/G (rs1610696) polymorphic sites, which were individually annotated according to previous reports [22, 49]. Haplotypes and genotypes were assigned and named, including for homozygous and heterozygous individuals.
Measuring sHLA-G plasma expression levels
A Bio Vendor (Oxford Biosystems Ltd. UK) sHLA-G ELISA kit was used for the detection of soluble forms of human leucocyte antigen-G (sHLA-G) in archived frozen serum samples from patients and healthy subjects. The assay was performed according to the manufacturer’s instructions. The reagents were prepared fresh at the time of assay and all reagents were brought to RT prior to use.
Peripheral Blood Mononuclear Cell (PBMC) isolation and stimulation with HIV-1 Env pseudo-typed virus particles</P>
HIV-1 pseudo-typed virus particles were generated composed of HIV-1 structural proteins and expressing an HIV1 CXCR4 envelope (Env) gp120/41 glycoprotein (CXCR4 tropic - LAI). We elected to utilize a CXCR4 using Env protein of HIV-1 as this is expressed at higher levels on CD4 lymphocytes isolated from blood and under the assumption that both receptors are likely to stimulate in a similar manner. This is a chimeric construct in which two plasmids (p8.19, a construct containing gag-Pol and pCSFLW, a construct with firefly luc under the control of the CMV promoter) were used as separate viral backbone vectors (1000ng and 1500ng respectively) in conjunction with a plasmid expressing HIV-1 Env LAI (900ng). 293T cells and PEI were used for plasmid transfection and media was changed 12-16 hours post-transfection with viral stocks harvested 48 hrs later. The harvested supernatant was added to peripheral blood mononuclear cells (PBMC) from healthy subjects and the supernatant containing the virus was taken. Lastly, the supernatants were harvested to different production volumes depending on experimental needs. PBMCs, prepared from healthy subjects by Ficoll density gradient separation, at a concentration of 1 x 106 /ml were cultured with and without the HIV-1 LAI construct for 7 days in a growth medium. Previous studies using CMV antigen stimulation had shown that HLA-G expression levels were shown to be modulated at day 7 post-exposure [2].
HLA-G mRNA expression levels determined through real-time PCR analysis
RNA was extracted from stimulated and unstimulated PBMC and converted into cDNA using a high capacity cDNA reverse transcription kit (Applied Biosystems). qPCR was then carried out using a TaqMan® probe covering exons 5 and 6 (Assay ID: Hs00365950_g1, Applied Biosystems Inc. USA). The master mix (TaqMan® Universal Master Mix II, with UNG, Applied Biosystems) and the control positive housekeeping gene was used as a control (Assay ID: Hs01087168_m1, Applied Biosystems). Assays were carried out in duplicate using cDNA from 9 stimulation experiments. Differences in HLA-G expression between unstimulated and stimulated cultures were calculated using the ΔCT method.
HLA-G cell surface expression levels determined through FACS analysis
PBMCs were analyzed at day 7 following culture with or without HIV-1 pseudo-typed virus particles were phenotyped with a panel of monoclonal antibodies (mAbs) corresponding to the antigens expressed by defined groups of cells: anti-CD3-FITC (IgG2a, no. 317306, Biolegend, London); anti-CD4-PE (IgG2b; no. 317410, Biolegend); anti-CD8-PE (IgG1; no. 9012-0087, eBioscience, Hatfield, UK); anti-CD19-PE (IgG1; no. 12-0198- 42, eBioscience); anti-CD56-PE (IgG1, no. 12-0567-42,eBioscience); anti-HLA-G-APC (IgG1; clone MEM-G/9, no. A15708, Life Technologies, Paisley, UK); isotype control-PE (IgG2b; no. ab91532, AbCam, Cambridge, UK); isotype control-APC (IgG1; MG105, Life technologies). The Abs were selected on the basis of optimal matching of three fluorochromes (FITC, PE, and APC), and 2- or 3-colour staining was performed using a BD Accuri C6 flow cytometer (BD Biosciences, Oxford). The main populations tested for changes in expression of HLA-G were CD4+, CD8+, and CD56+ T cells, B cells, and NK cells. The mean proportions of each subset of lymphocytes expressing HLA-G were measured on cells with or without culture with HIV-Env pseudo-typed virus particles for 7 days. Results were expressed as mean % HLA-G+ cells for different leucocyte subsets after subtracting background staining with an appropriate isotype control antibody
Statistical analysis
Two-way ANOVA and χ2 tests were used to analyze differences between genotype and allele frequencies between patient groups and controls. For analysis of soluble HLA-G levels, a Mann-Whitney U-test was used as the data were not normally distributed. A value of P<0.05 was regarded as statistically significant.
Results
HLA-G 3’ UTR genotypes associated with HRSN status
HLA-G 3’ UTR variant genotypes and haplotypes were determined for three defined clinical groups from the ACS along
with controls (n=275 total): HIV-1+ MSM (n=133), HIV-1+ IDU (n=50), HRSN (n=32) along with control subjects (n=85). The alleles identified included: the eight previously reported HLA-G 3’UTR polymorphic sites [14-bp INS/DEL (rs1704), +3003 C/T (rs1707), +3010C/G (rs1710), +3027A/C (rs17179101), +3035 C/T (rs17179108), +3142 C/G (rs1063320), +3187 A/G (rs9380142) and +3196 C/G (rs1610696)] [25-27] with no other polymorphic sites observed in this region. The rationale was to identify whether frequencies of any of the high or low expressing alleles differed between patient groups, in particular, the HRSN group, which differed functionally from the other patient groups in response to virus exposure. The genotype frequencies of the eight HLA-G 3’UTR polymorphic sites observed within these patient groups were determined (Table 1). Upon performing a χ2 test to assess the occurrence of these SNPs among the four designated patient groups, significant differences were found (Table 1). The data were also evaluated at the level of individual alleles (Table 2) and again significant differences were observed. Both the 3035T and 3010C alleles are associated with low levels of HLA-G expression and proportions of these were significantly greater in the HRSN group than the other HIV+ groups (MSM and IDU) as well as the control individuals (Table 2). Additionally, the 3003C/T and 14bpI/D dimorphisms showed significant differences from the Hardy-Weinberg Equilibrium (Table 1).
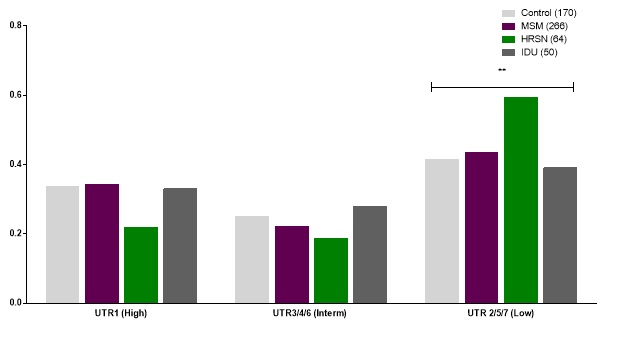
The HLA-G 3’UTR haplotypes were then allocated for each patient group and healthy controls. Patients were allocated into the seven major haplotypes UTR1-7 [27]. Based upon the alleles associated with high or low levels of HLA-G expression, haplotypes were allocated into one of three groups, potentially giving high (UTR1), intermediate (UTRs 3, 4 & 6) or low levels of HLA-G expression (UTRs 2, 5 & 7). When haplotype groups for each patient group and controls were analyzed, MSM and IDU groups were similar to controls but there were significantly higher proportions of low expressing haplotypes in the HRSN group, along with lower proportions of the high expressing haplotype UTR1 (Figure 1).
Serum sHLA-G levels do not differ between HIV+, HRSN, and control individuals
As a measure of HLA-G expression, sHLA-G serum levels were assayed, where sufficient sample was available, in HIV-1 groups [HIV+ (n=27) and HRSN (n=20)] and compared to healthy control subjects (n=20). Control sera provided a mean level of 35.5+/-32 u/ml, MSM gave 100+/-69 u/ml (n=20) and HRSN gave 116.1+/-79 u/ml (n=20). When compared no significant differences between sHLA-G levels between the HIV-1 groups (HIV+ and HRSN) or with controls were found (Mann-Whitney U-test; P>0.05).
HLA-G cell-surface expression does not change on immune cells when exposed to HIV-1 in vitro
Since HLA-G 3’ UTR genetic polymorphisms associate with HIV-1 HRSN individuals, which are known to be linked to lower HLA-G expression, we aimed to identify whether exposure to the virus results in lower HLA-G expression. We developed an assay where PBMC from healthy subjects were exposed to a high concentration of infectious but non-replicative competent HIV1. The reasoning being that HRSN individuals have previously been exposed to either infectious HIV-1, defective viral particles, or viral antigens at mucosal surfaces. We utilized a pseudo-typed virus system where HIV-1 backbone plasmids are co-transfected into a producer cell-line along with a plasmid expressing the HIV-1 LAI Env protein (CXCR4 utilizing). This virus undergoes entry and integration but cannot subsequently replicate. PBMC from a panel of 11 healthy subjects independent of their UTR genotype were stimulated for 7 days in vitro with the HIV-1 LAI Env expressing pseudo-typed virus particles and where unstimulated cells served as a control. Different leucocyte populations were assessed for changes in HLA-G cell-surface expression by flow cytometry.
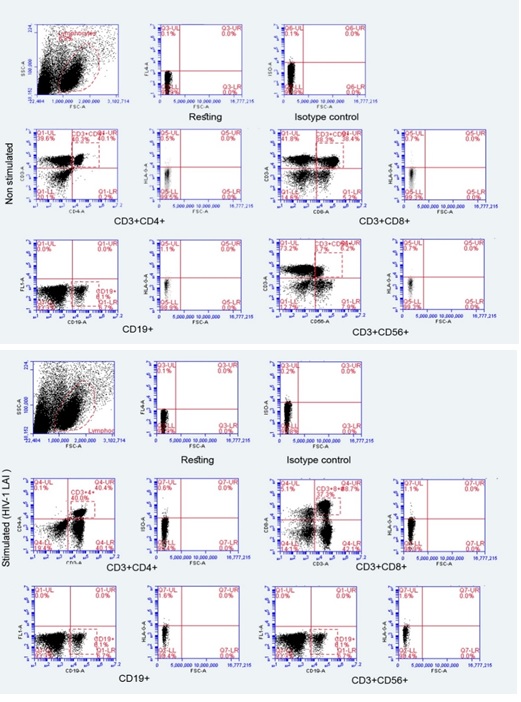
Representative scatter plots are shown (Figure 2) and comparisons of the proportions of HLA-G+ cells on various cellular subsets stimulated with HIV-1 pseudo-typed viruses were made. Slight increases in the proportions of different leucocyte subpopulations expressing HLA-G following stimulation were observed. CD3+CD4+ lymphocytes were revealed to have the highest increase in the proportion of HLA-G expressing cells, followed by CD3+CD8+ cells, followed by CD19+ cells, although all differences were < 10% and none of them were statistically significant (Figure 2). The remaining subsets of PBMC showed no differences between stimulated and unstimulated cells in all of the 11 subjects.
HLA-G mRNA expression levels were measured using real-time PCR and utilizing mRNA isolated from PBMCs that had been stimulated with HIV-1 LAI pseudo-typed virus particles and compared to unstimulated cells. The ΔCT values using an HLA-G probe for unstimulated and stimulated cells were 39.88 and 40.59 respectively (n=9 subjects), but these differences were not statistically significant (data not are shown).
Discussion
HIV-1 infection and viral escape processes have been extensively studied [17]. Although an apparently appropriate immune response is initially induced by HIV-1 it typically evades the immune responses mounted [4]. Several studies have suggested that there is an increased expression of HLA-G after HIV1 infection and that the immunomodulatory molecule HLA-G is capable of suppressing immune responses, although the real involvement of this molecule in virus infection and immune induction remain elusive [50]. In the present study, we investigated the association between the 14 bp INS/DEL and other HLA-G 3’UTR gene polymorphisms with HIV-1 infection. The evaluations were performed using different HIV-1 groupings from the ACS (MSM HIV-1+, IDU HIV-1+, and HRSN) and utilizing regionally-matched controls. We observed an increased frequency of the genotypes related to low HLA-G expression (UTR-2, 5, and 7) in HRSN individuals in comparison to MSM HIV-1+, IDU HIV-1+or healthy uninfected individuals. Taken together this suggests that low HLA-G expression alleles may play a role in protecting against horizontal HIV-1transmission.
Previous studies have linked HLA-G genotypic variation with susceptibility to HIV-1[32, 35, 51] with low levels of expression being protective [35]. Investigations of the 14bp Ins/Del dimorphism have been inconclusive, with some studies identifying the insertion allele as protective against HIV-1 infection or vertical transmission [26, 46], while others showed the deletion allele to be protective [13, 16, 34, 44]. The suggestion has been made that the 14bp INS/DEL dimorphism is not directly related to HIV-1 susceptibility [1] and it has been proposed that genetic variations in levels of expression are more relevant to HIV-1 vertical transmission in view of the extensive-expression of HLA-G at the materno foetal interface [45]. Here we found no difference in the frequencies of UTR genotypes in individuals either infected via MSM or IDU transmission and no difference between these two groups and control individuals.
The 14bp INS/DEL dimorphism is only one of several 3’ UTR polymorphisms and consideration of genetic variation across the whole region is more likely to give an indication of the potential levels of HLA-G expression. The HLA-G 3′UTR region has a number of binding sites for miRNAs and the C/G polymorphism at position +3142 is the putative binding site for at least three miRNAs: hsa-mir-148a, hsa-mir-148b, and hsamir152, and has been subsequently confirmed to be the binding site for mir152 and it is noted that this binding reduces HLA-G expression [7, 57]. However, the effect of this polymorphism in miRNA binding has been recently questioned as although mir152 and mir-148a were found to bind to HLA-G 3′UTR and down regulate its mRNA, the C/G polymorphism at +3142 was found to have no effect on miRNA binding and efficacy [33]. The +3187G allele, associated with increased HLA-G expression and characteristic of the UTR1 haplotype, was related to increased vertical transmission of HIV in a South African population [19]. Although the present results suggest that low-expressing HLA-G UTR haplotypes had a protective effect against horizontal HIV-1 transmission, we acknowledge that our findings are restricted to patients from the Amsterdam Cohort, who are predominantly of Caucasian origin. Different findings have been reported for African populations [1, 26] and further genetic studies are warranted in different populations.
Serum levels of sHLA-G were reported to be significantly elevated in HIV-infected subjects [18] and sHLA-G has the ability to inhibit T lymphocyte proliferation and NK cell function [11, 38, 48]. sHLA-G could potentially interact with one of its ligands, KIR2DL4, found on NK cells and other PBMCs in order to moderate inflammatory mediator synthesis. In addition, the binding of sHLA-G with ILT-4 on DCs can result in activating T reg cells which in turn participate in the induction of tolerance favouring virus evasion [27]. There have also been reports of HLA-G-expressing regulatory T cells to be found in HIV-1 infected individuals [28, 30] which are potential mediators of immune suppression of anti-HIV cellular immune responses. In the current study, sHLA-G serum levels did not differ significantly between the HIV-1 groups and controls. This may indicate that the sHLA-G protein is not the main mediator of immune suppression in HIV-1 infected individuals, but rather that cell plasma membrane-associated HLA-G is more important functionally. Alternatively, most sHLA-G may be taken up by cell surface HLA-G receptors and patterns of expression of the known receptors KIR2DL4, ILT-2, and ILT-4, may be relevant and may potentially vary in the presence of HIV infection.
Changes in HLA-G expression were explored in response to HIV-1 LAI pseudo-typed virus in PBMC from healthy subjects. The proportions of HLA-G+ cells in various cellular populations harvested on Day 7 of culture with HIV Ag were compared to resting proportions of HLA-G+ cells. CD3+CD4+, CD3+CD8+, and CD3+CD56+ lymphocytes revealed slight but not statistically significant elevations of HLA-G expression on Day 7. Numerous studies were aimed at observing changes in the expression of HLA-G in the early stage of infection by HIV1 and through disease progression. During HIV-1 infection the HLA-G1 isoform was found to be down regulated on macrophages [14] but in an earlier report, both monocytes and T cells were found to express HLA-G in HIV+ patients [29]. HLA-G+ CD8+ cytotoxic T cells were reported to be associated with protection against HIV-1 [53]. In the present experiments, not all HIV genes were present, only structural proteins as well as Env, therefore it remains possible that other HIV-1 protein (such as Nef) may be responsible for the modulation of HLA-G expression [42].
In conclusion, we identify that genetic polymorphisms within the 3’ UTR region (14 bp INS/DEL) of the HLA-G gene associate with HRSN individuals from the ACS and which differ from HIV-1+ (MSM and IDU) or control individuals. The main finding is that an increased frequency of UTR genotypes related to low HLA-G expression (UTR-2, 5, and 7) was found in HRSN as compared to other patient groups and uninfected controls. This suggests that low expressing HLA-G alleles may play a role in protecting against HIV-1 infection. It would be of great interest to extend these studies to other populations and to determine whether HLA-G expression in HIV-1 infected individuals can be linked to other genetic polymorphisms and associate with disease outcome or treatment success.
Acknowledgments
The authors declare that there are no conflicts of interest.
AA conducted the experiments and analyzed the results; LM provided the HIV vector and advised on antigen stimulation experiments; WAP, GP, and BFF advised on experimental design and analysis of results; LB provided statistical advice; NK provided samples and advised on interpretation of results; SEC coordinated the study and wrote the manuscript and all co-authors read and commented on the manuscript. Dr. Andrea Jorgensen (Department of Biostatistics, Institute of Translational Medicine, University of Liverpool) advised on the statistical methods.
- Aghafar MZ, Witt C, Kamarulzaman A, et al. (2012) Genetic variations in loci relevant to natural killer cell function are affected by ethnicity but are generally not correlated with susceptibility to HIV-1.Tissue Antigens 79: 367-371.
- Albayati Z, Alyami A, Alomar S, et al. (2017) The Influence of Cytomegalovirus on Expression of HLA-G and its Ligand KIR2DL4 by Human Peripheral Blood Leucocyte Subsets. Scand J Immunol 86: 396-407.
- Amiot L, Vu N, Rauch M, et al. (2014) Expression of HLA-G by mast cells is associated with hepatitis C virus-induced liver fibrosis. J Hepatol 60: 245-252.
- Boutwell CL, Rolland MM, Herbeck JT, et al. (2010) Viral evolution and escape during acute HIV-1 infection. J Infect Dis 202 Suppl 2: S309-314.
- Cabello A, Rivero A, Garcia MJ, et al. (2003) HAART induces the expression of HLA-G on peripheral monocytes in HIV-1 infected individuals. Hum Immunol 64: 1045-1049.
- Castelli EC, Mendes-Junior CT, Deghaide NH, et al. (2010) The genetic structure of the 3'untranslated region of the HLA-G gene: polymorphisms and haplotypes. Genes Immun 11: 134-141.
- Castelli EC, Moreau P, Oya e Chiromatzo A, et al. (2009) In silico analysis of microRNAS targeting the HLA-G 3' untranslated region alleles and haplotypes. Hum Immunol 70: 1020-1025.
- Castelli EC, Veiga-Castelli LC, Yaghi L, et al. (2014) Transcriptional and posttranscriptional regulations of the HLAG gene.J Immunol Res 2014: 734068.
- Celsi F, Catamo E, Kleiner G, et al. (2013) HLA-G/C, miRNAs, and Their Role in HIV Infection and Replication. BioMed Res Intl 2013: 693643.
- Chen HX, Chen BG, Shi WW, et al. (2011) Induction of cell surface human leukocyte antigen-G expression in pandemic H1N1 2009 and seasonal H1N1 influenza virus-infected patients. Hum Immunol 72: 159-165.
- Contini P, Ghio M, Poggi A, et al. (2003) Soluble HLAA,-B,-C and -G molecules induce apoptosis in T and NK CD8+ cells and inhibit cytotoxic T cell activity through CD8 ligation. Eur J Immunol 33: 125-134.
- Coutinho RA. (1998) The Amsterdam Cohort Studies on HIV infection and AIDS. J Acquir Immune DeficSyndr Hum Retrovirol 17 Suppl 1: S4-8.
- da Silva GK, Vianna P, Veit TD, et al. (2014) Influence of HLA-G polymorphisms in human immunodeficiency virus infection and hepatitis C virus co-infection in Brazilian and Italian individuals. Infect Genet Evol 21: 418-423.
- Derrien M, Pizzato N, Dolcini G, et al. (2004) Human immunodeficiency virus 1 down regulates cell surface expression of the non-classical major histocompatibility class I molecule HLA-G1.J Gen Virol 85: 1945-1954.
- Donaghy L, Gros F, Amiot L, et al. (2007) Elevated levels of soluble non-classical major histocompatibility class I molecule human leucocyte antigen (HLA)-G in the blood of HIVinfected patients with or without visceral leishmaniasis. Clin Exp Immunol 147: 236-240.
- Fabris A, Catamo E, Segat L, et al. (2009) Association between HLA-G 3'UTR 14-bp polymorphism and HIV vertical transmission in Brazilian children. AIDS 23: 177-182.
- Fanales-Belasio E, Raimondo M, Suligoi B, Butto S. (2010) HIV virology and pathogenetic mechanisms of infection: a brief overview. Ann Ist Super Sanita 46: 5-14.
- German Advisory Committee Blood SAoPTbB. (2016) Human Immunodeficiency Virus (HIV). Transfusion Medicine and Hemotherapy 43: 203-222.
- Hong HA, Paximadis M, Gray GE, et al. (2015) Maternal human leukocyte antigen-G (HLA-G) genetic variants associate within utero mother-to-child transmission of HIV-1 in Black South Africans. Infect Genet Evol 30: 147-158.
- Huang J, Burke P, Yang Y, et al. (2010) Soluble HLAG inhibits myeloid dendritic cell function in HIV-1 infection by interacting with leukocyte immunoglobulin-like receptor B2. J Virol 84: 10784-10791.
- Huang M-B, Ye L, Liang B-Y, Ning C-Y, Roth WW, Jiang J-J, et al. (2016) Characterizing the HIV/AIDS Epidemic in the United States and China. Int J Environ Res Public Health 13: 30.
- Hviid TV, Christiansen OB (2005) Linkage disequilibrium between human leukocyte antigen (HLA) class II and HLA-G – possible implications for human reproduction and autoimmune disease. Hum Immunol 66: 688–699.
- Lajoie J, Fontaine J, Tremblay C, et al. (2009) Persistence of high levels of blood soluble human leukocyte antigen-G is associated with rapid progression of HIV infection. AIDS 23: 437- 1440.
- Lajoie J, Hargrove J, Zijenah LS, et al. (2006) Genetic variants in nonclassical major histocompatibility complex class I human leukocyte antigen (HLA)-E and HLA-G molecules are associated with susceptibility to the heterosexual acquisition of HIV-1. J Infect Dis 193: 298-301.
- Lajoie J, Massinga Loembe M, Poudrier J, et al. (2010) Blood soluble human leukocyte antigen G levels are associated with human immunodeficiency virus type 1 infection in Beninese commercial sex workers. Hum Immunol 71: 182-185.
- Larsen MH, Zinyama R, Kallestrup P, et al. (2013) HLAG 3' untranslated region 14-base pair deletion: association with poor survival in an HIV-1-infected Zimbabwean population. J Infect Dis 207: 903-906.
- LeMaoult J, Krawice-Radanne I, Dausset J, Carosella ED (2004) HLA-G1-expressing antigen-presenting cells induce immunosuppressive CD4+ T cells. Proc Natl Acad Sci U S A 101: 7064-7069.
- Li C, Toth I, Schulze ZurWiesch J, et al. (2013) Functional characterization of HLA-G⁺ regulatory T cells in HIV-1 infection. PLoS Pathog 9: e1003140.
- Lozano JM, González R, Kindelán JM, et al. (2002) Monocytes and T lymphocytes in HIV-1-positive patients express HLA-G molecule. AIDS 16: 347-351.
- Lozano JM, González R, Luque J, et al. (2009) CD8(+) HLA-G(+) regulatory T cells are expanded in HIV-1-infected patients. Viral Immunol 22: 463-465.
- Lucena-Silva N, Monteiro AR, de Albuquerque RS, et al. (2012) Haplotype frequencies based on eight polymorphic sites at the 3' untranslated region of the HLA-G gene in individuals from two different geographical regions of Brazil. Tissue Antigens 79: 272–278.
- Luo M, Czarnecki C, Ramdahin S, et al. (2013) HLA-G and mother-child perinatal HIV transmission. Hum Immunol 74: 459-463.
- Manaster I, Goldman-Wohl D, Greenfield C, et al. (2012) MiRNA-mediated control of HLA-G expression and function. PloS One 7: e33395.
- Martelli-Palomino G, Pancotto JA, et al. (2013) Polymorphic sites at the 3' untranslated region of the HLA-G gene are associated with differential hla-g soluble levels in the Brazilian and French population. PLoS One 8: e71742.
- Matte C, Lajoie J, Lacaille J, et al. (2004) Functionally active HLA-G polymorphisms are associated with the risk of heterosexual HIV-1 infection in African women. AIDS 18: 427-431.
- Menier C, Rouas-Freiss N, Favier B, et al. (2010) Recent advances on the non-classical major histocompatibility complex class I HLA-G molecule. Tissue Antigens 75: 201-206.
- Moodley S, Bobat R (2011) Expression of HLA-G1 at the placental interface of HIV-1 infected pregnant women and vertical transmission of HIV. Placenta 32: 778-782.
- Morandi F, Pistoia V (2013) Soluble HLA-G modulates miRNA-210 and miRNA-451 expression in activated CD4+ T lymphocytes. Int Immunol 25: 279-285.
- Morandi F, Rizzo R, Fainardi E, et al. (2016) Recent Advances in Our Understanding of HLA-G Biology: Lessons from a Wide Spectrum of human diseases. J Immunol Res 4326495.
- Murdaca G, Contini P, Setti M, et al. (2009) Behavior of non-classical soluble HLA class G antigens in human immunodeficiency virus 1-infected patients before and after HAART: comparison with classical soluble HLA-A, -B, -C antigens and potential role in immune-reconstitution. Clin Immunol 133: 238-244.
- Murdaca G, Contini P, Setti M, et al. (2011) Soluble human leukocyte antigen-G serum levels in patients with acquired immune deficiency syndrome affected by different disease-defining conditions before and after antiretroviral treatment. Hum Immunol 72: 712-716.
- Pizzato N, Derrien M, Lenfant F (2004) The short cytoplasmic tail of HLA-G determines its resistance to HIV-1 Nefmediated cell surface downregulation. Hum Immunol 65: 1389- 1396.
- Rizzo R, Gabrielli L, Bortolotti D, et al. (2016) Study of Soluble HLA-G in Congenital Human Cytomegalovirus Infection. J Immunol Res 3890306.
- Segat L, Catamo E, Fabris A, et al. (2009) HLA-G 3' UTR haplotypes and HIV vertical transmission. AIDS 23: 1916- 1918.
- Segat L, Crovella S (2012) HLA-G 14bp del/ins genetic variation: association with susceptibility to human immunodeficiency virus-1 vertical transmission but not with human immunodeficiency virus-1 infection through horizontal transmission. Tissue Antigens 80: 12-13
- Segat L, Zupin L, Kim HY, et al. (2014) HLA-G 14 bp deletion /insertion polymorphism and mother-to-child transmission of HIV. Tissue Antigens 83: 161-167.
- Shi WW, Lin A, Xu DP, et al. (2011) Plasma soluble human leukocyte antigen-G expression is a potential clinical biomarker in patients with hepatitis B virus infection. Hum Immunol 72: 1068-1073.
- Spaggiari GM, Contini P, Carosio R, et al. (2002) Soluble HLA class I molecules induce natural killer cell apoptosis through the engagement of CD8: evidence for a negative regulation exerted by members of the inhibitory receptor superfamily. Blood 99: 1706-1714.
- Thibodeau V, Lajoie J, Labbé AC, et al. (2011) High level of soluble HLA-G in the female genital tract of Beninese commercial sex workers is associated with HIV-1 infection. PLoS One 6: e25185.
- Tripathi P, Agrawal S (2007) The role of human leukocyte antigen E and G in HIV infection. AIDS 21: 1395-1404.
- Turk WJ, Kimani J, Bielawny T, et al. (2013) Associations of human leukocyte antigen-G with resistance and susceptibility to HIV-1 infection in the Pumwani sex worker cohort. AIDS 27: 7-15.
- Veit TD, Chies JA (2009) Tolerance versus immune response – microRNAs as important elements in the regulation of the HLA-G gene expression. Transpl Immunol 20: 229–231.
- Viganò S, Negrón JJ, Tse S, et al. (2017) HLA-G+ HIV1-specific CD8+ T cells are associated with HIV-1 immune control. AIDS 31: 207-212.
- Weng PJ, Fu YM, Ding SX, et al. (2011) Elevation of plasma soluble human leukocyte antigen-G in patients with chronic hepatitis C virus infection. Hum Immunol 72: 406-411.
- Yie SM, Li LH, Xiao R, Librach CL (2008) A single basepair mutation in the 3’-untranslated region of HLA-G mRNA is associated with pre-eclampsia. Mol Hum Reprod 14: 649–653.
- Zheng XQ, Chen XQ, Gao Y, et al. (2014) Elevation of human leukocyte antigen-G expression is associated with the severe encephalitis associated with neurogenic pulmonary edema caused by Enterovirus 71. Clin Exp Med 14: 161-167.
- Zhu XM, Han T, Wang XH, et al. (2010) Over expression of miR-152 leads to reduced expression of human leukocyte antigen-G and increased natural killer cell-mediated cytolysis in JEG-3 cells. Amer J Obstet Gynecol 202: 592.e1-7.

Tables at a glance